Cellular membranes are made up of lipid bilayers that can be highly curved, an essential feature critical for processes such as endocytosis, organelle synthesis, cell division, and intracellular transport. Current understanding suggests that specialized proteins are responsible for membrane curvature through two mechanisms (Figure 1). The first is membrane scaffolding by proteins or complexes that assemble into a curved shape. These scaffolding proteins can bend membranes using anchors that cause the membrane to follow the contour of the protein. Proteins such as amphiphysin and dynamin are thought to act as scaffolds that form thin membrane necks prior to separation of the membrane regions (fission). The second mechanism thought responsible for membrane bending involves proteins with wedge-like helices that contains parts that are amphipathic, meaning they are both oil-loving (lipophilic) and water-loving (hydrophilic). These helices partially insert themselves into the lipophilic membrane and push the lipid molecules apart. Epsin1 is an example of such a protein.

However, some computational studies have cast doubt on this mechanism, predicting that to generate the observed curvature, proteins must cover nearly 100% of the membrane surface. Such a situation is physiologically unlikely.
CRF researchers Carl Hayden, Darryl Sasaki and Jeanne Stachowiak (now at the University of Texas, Austin), along with collaborators Eva Schmid and Daniel Fletcher at the University of California Berkeley, and others, have studied this problem with a variety of methods and proposed a third mechanism for bending cellular membranes—protein-protein crowding.
Membrane curvature and tubulation: Fluorescent confocal microscopy
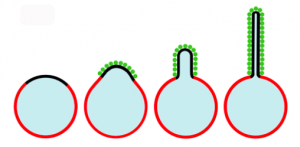
Inspired by the recent discovery that cells transport materials from one cell to another using lipid nanotubes, the CRF researchers first studied how it might be possible to create such nanotubules synthetically. The group initially used Laboratory Directed Research and Development (LDRD) funds for their work. In work published in a 2010 article in the Proceedings of National Academy of Sciences, studies of protein-lipid interactions in giant lipid vesicles containing domains with a high affinity for poly-histadine (his-tagged) proteins were reported. Experiments with green fluorescent protein (GFP) showed that his-tagged proteins crowded together densely in the high protein affinity domains, bending them into lipid buds and tubules. No protein insertion or scaffolding was needed to bend the membrane. Further, the formation of the tubules was dependent on both the presence of high protein affinity lipids and a high density of protein attachment. Although this was a simplified model, the results implied that confinement of a high surface density of proteins to lipid domains could cause the formation of tubules. The protein crowding membrane curvature mechanism is illustrated in Figure 2.
Studies of epsin1: Fluorescence confocal microscopy
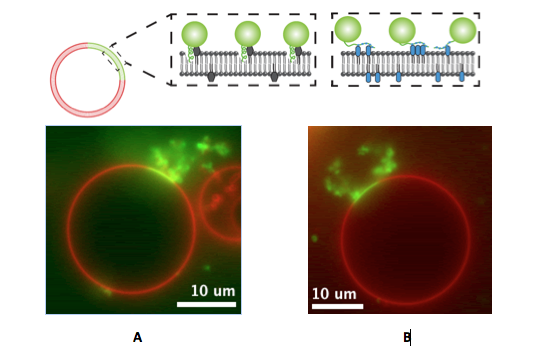
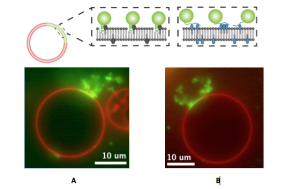
Studies with proteins thought to cause membrane bending in cells were then undertaken to investigate whether protein crowding might play a role in actual biological systems. This work came about through a collaboration between the CRF researchers with chemistry and materials expertise and the UC Berkeley scientists experienced in biochemistry. In order to compare the amphipathic helix insertion mechanism to membrane bending by protein crowding, the team investigated epsin1, a protein important in endocytosis. Epsin1 has a membrane binding N-terminal homology (ENTH) domain, a part of a protein that can function and exist independent of the rest of the protein chain. A donor fluorescence decay-based Förster resonance energy transfer (FRET) method was used with a confocal microscope to make in situ density measurements of proteins on complex structured membranes (again on artificial membrane vesicles). The wild type ENTH domain, which inserted an amphipathic helix upon binding, was compared with a mutant ENTH domain in which the helix had been replaced with an engineered his-tag to enable binding to the membrane. Both the wild type and the mutant proteins were found to bend the membrane at similar densities, indicating that helix insertion was not necessary (Figure 3). The measurements further indicated that the lateral pressure generated by protein surface coverage greater than~20% is sufficient to bend membranes.
This work was published in 2012 in nature cell biology (online), and topped the most-viewed list of articles in the month of its publication.
What does this mean?
Unsurprisingly, the work described here has some controversial implications that can change the way scientists understand membrane curvature. Although this work so far has been conducted in in vitro systems, the results indicate that further study in more intricate systems under biologically relevant conditions is warranted. Says Hayden, “We started out studying how to artificially make lipid nanotubules like those seen between cells and ended up using what we learned to investigate the details of a potential new mechanism for membrane bending in cells. In a sense, we’ve come full circle.” Wherever further studies lead the team, it’s clear that collaborations between researchers in widely different fields can result in significant, and unexpected, insights that benefit a broad community.
This work was supported by the US Department of Energy, Office of Basic Energy Sciences, Division of Materials Science and Engineering and Division of Chemical Sciences, Geosciences, and Biosciences; the Laboratory Directed Research and Development program at Sandia National Laboratories; the NIH NIGMS and Nanomedicine Development Centers; a Sealy and Smith Foundation grant to the Sealy Center for Structural Biology and Molecular Biophysics; and the Miller Institute for Basic Research in Science.